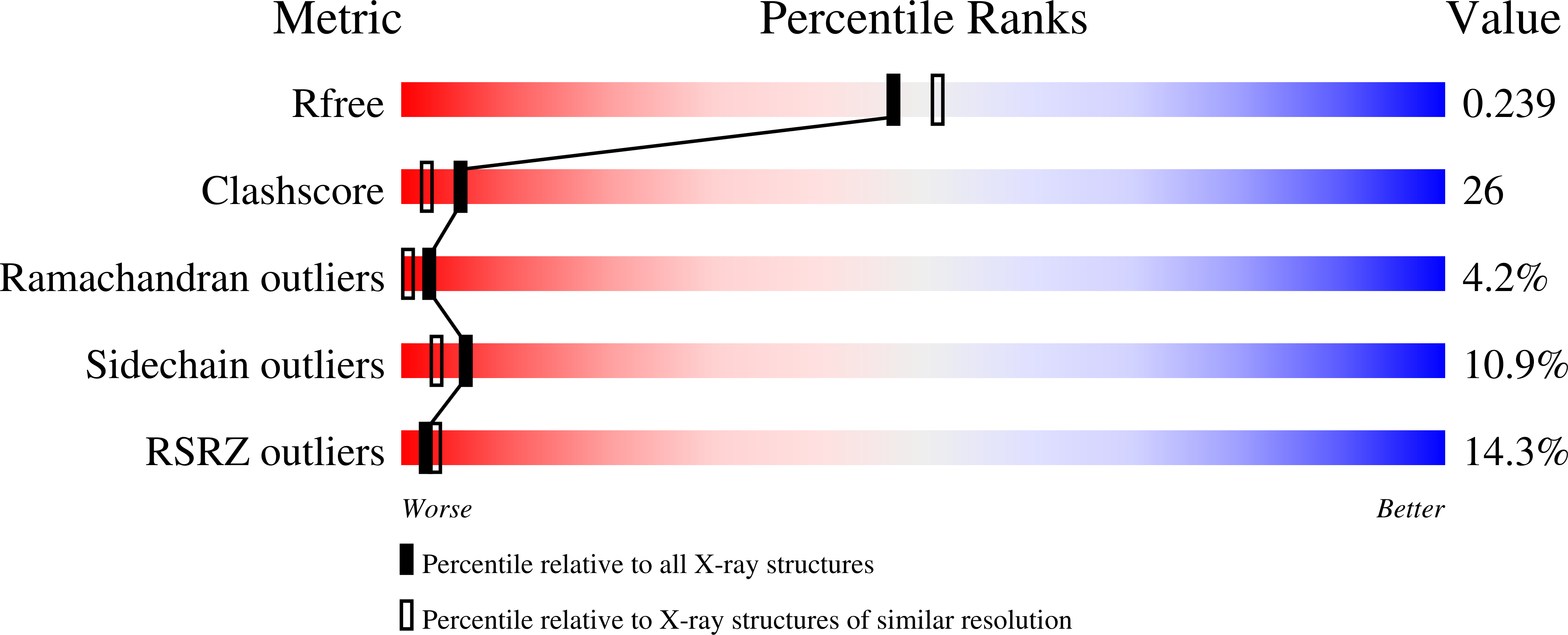
Crystal structure of Escherichia coli MazG, the regulator of nutritional stress response.
Lee, S., Kim, M.H., Kang, B.S., Kim, J.S., Kim, G.H., Kim, Y.G., Kim, K.J.(2008) J Biol Chem 283: 15232-15240
- PubMed: 18353782
- DOI: https://doi.org/10.1074/jbc.M800479200
- Primary Citation of Related Structures:
3CRA, 3CRC - PubMed Abstract:
MazG is a nucleoside triphosphate pyrophosphohydrolase that hydrolyzes all canonical nucleoside triphosphates. The mazG gene located downstream from the chromosomal mazEF "addiction module," that mediated programmed cell death in Escherichia coli. MazG activity is inhibited by the MazEF complex both in vivo and in vitro. Enzymatic activity of MazG in vivo affects the cellular level of guanosine 3',5'-bispyrophosphate (ppGpp), synthesized by RelA under amino acid starvation. The reduction of ppGpp, caused by MazG, may extend the period of cell survival under nutritional stress. Here we describe the first crystal structure of active MazG from E. coli, which is composed of two similarly folded globular domains in tandem. Among the two putative catalytic domains, only the C-terminal domain has well ordered active sites and exhibits an NTPase activity. The MazG-ATP complex structure and subsequent mutagenesis studies explain the peculiar active site environment accommodating all eight canonical NTPs as substrates. In vivo nutrient starvation experiments show that the C terminus NTPase activity is responsible for the regulation of bacterial cell survival under nutritional stress.
Organizational Affiliation:
Pohang Accelerator Laboratory, Pohang University of Science and Technology, Pohang, Kyungbuk, Korea.