Effects of Synergistic Inhibition on α-glucosidase by Phytoalexins in Soybeans
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Inhibition of α-glucosidase
2.3. Enzyme Kinetics for α-glucosidase
- (Competitive inhibition)
- (Uncompetitive inhibition)
- (Noncompetitive inhibition)
- (Mixed inhibition)
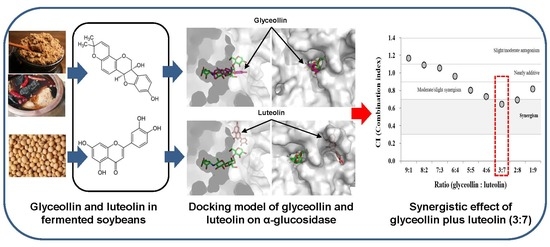
2.4. Docking Studies
2.5. Synergistic Effects on α-glucosidase Inhibition
2.6. Determination of the Combination Index (CI)
2.7. Statistical Analysis
3. Results
3.1. Potential of Phytoalexins Derived from Soybeans to Inhibit α-Glucosidase
3.2. Determination of Inhibition Modes and Ki Values on Polyphenols Derived from Soybeans on α-glucosidase
3.3. Structural Bases for the Modes of Inhibition
3.4. Combined Effects of Glyceollin Plus Luteolin on α-glucosidase Inhibition
3.5. Confirmation of Synergism by CI Equation with Glyceollin Plus Luteolin
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hu, C.; Wong, W.T.; Wu, R.; Lai, W.F. Biochemistry and use of soybean isoflavones in functional food development. Crit. Rev. Food Sci. Nutr. 2019, 59, 1–15. [Google Scholar] [CrossRef]
- Rizzo, G.; Baroni, L. Soy, soy foods and their role in vegetarian diets. Nutrients 2018, 10, 43. [Google Scholar] [CrossRef] [Green Version]
- Nagata, C.; Mizoue, T.; Tanaka, K.; Tsuji, I.; Tamakoshi, A.; Matsuo, K.; Wakai, K.; Inoue, M.; Tsugane, S.; Sasazuki, S. Soy intake and breast cancer risk: An evaluation based on a systematic review of epidemiologic evidence among the Japanese population. Jpn. J. Clin. Oncol. 2014, 44, 282–295. [Google Scholar] [CrossRef] [Green Version]
- Roblet, C.; Doyen, A.; Amiot, J.; Pilon, G.; Marette, A.; Bazinet, L. Enhancement of glucose uptake in muscular cell by soybean charged peptides isolated by electrodialysis with ultrafiltration membranes (EDUF): Activation of the AMPK pathway. Food Chem. 2014, 147, 124–130. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Zhang, X.; Xiang, Y.B.; Yang, G.; Li, H.; Fazio, S.; Linton, M.; Cai, Q.; Zheng, W.; Gao, Y.T.; et al. Association of soy food intake with risk and biomarkers of coronary heart disease in Chinese men. Int. J. Cardiol. 2014, 172, e285–e287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boué, S.M.; Isakova, I.A.; Burow, M.E.; Cao, H.; Bhatnagar, D.; Sarver, J.G.; Shinde, K.V.; Erhardt, P.W.; Heiman, M.L. Glyceollins, soy isoflavone phytoalexins, improve oral glucose disposal by stimulating glucose uptake. J. Agric. Food Chem. 2012, 60, 6376–6382. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Song, L.; Lee, Y.K.; Huang, D. The effects of fungal stress on the antioxidant contents of black soybeans under germination. J. Agric. Food Chem. 2010, 58, 12491–12496. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.S.; Jung, U.J.; Yeo, J.; Kim, M.J.; Lee, M.K. Genistein and daidzein prevent diabetes onset by elevating insulin level and altering hepatic gluconeogenic and lipogenic enzyme activities in non-obese diabetic (NOD) mice. Diabetes Metab. Res. Rev. 2008, 24, 74–81. [Google Scholar] [CrossRef] [PubMed]
- Scuro, L.S.; Simioni, P.U.; Grabriel, D.L.; Saviani, E.E.; Modolo, L.V.; Tamashiro, W.M.; Salgado, I. Suppression of nitric oxide production in mouse macrophages by soybean flavonoids accumulated in response to nitroprusside and fungal elicitation. BMC Biochem. 2004, 5, 5. [Google Scholar] [CrossRef] [Green Version]
- Palacios-Gonzalez, B.; Flores-Galicia, I.; Noriega, L.G.; Alemán-Escondrillas, G.; Zariñan, T.; Ulloa-Aguirre, A.; Torres, N.; Tovar, A.R. Genistein stimulates fatty acid oxidation in a leptin receptor-independent manner through the JAK2-mediated phosphorylation and activation of AMPK in skeletal muscle. Biochim. Biophys. Acta 2014, 1841, 132–140. [Google Scholar] [CrossRef]
- Yoon, E.K.; Kim, H.K.; Cui, S.; Kim, Y.H.; Lee, S.-H. Soybean glyceollins mitigate inducible nitric oxide synthase and cyclooxygenase-2 expression levels via suppression of the NF-κB signaling pathway in RAW 264.7 cells. Int. J. Mol. Med. 2012, 29, 711–717. [Google Scholar] [CrossRef]
- Lee, S.-H.; Lee, J.; Jung, M.H.; Lee, Y.M. Glyceollins, a novel class of soy phytoalexins, inhibit angiogenesis by blocking the VEGF and bFGF signaling pathways. Mol. Nutr. Food Res. 2013, 57, 225–234. [Google Scholar] [CrossRef]
- Kim, H.J.; Suh, H.J.; Kim, J.H.; Park, S.; Joo, Y.C.; Kim, J.S. Antioxidant activity of glyceollins derived from soybean elicited with Aspergillus sojae. J. Agric. Food Chem. 2010, 58, 11633–11638. [Google Scholar] [CrossRef]
- Lee, Y.S.; Kim, H.K.; Lee, K.J.; Jeon, H.W.; Cui, S.; Lee, Y.M.; Moon, B.J.; Kim, Y.H.; Lee, Y.S. Inhibitory effect of glyceollin isolated from soybean against melanogenesis in B16 melanoma cells. BMB Rep. 2010, 43, 461–467. [Google Scholar] [CrossRef]
- Chiba, S. Molecular mechanism in α-glucosidase and glucoamylase. Biosci. Biotechnol. Biochem. 1997, 61, 1233–1239. [Google Scholar] [CrossRef]
- Larner, J.; Lardy, H.; Myrback, K. Other glucosidases. In The Enzymes 4, 2nd ed.; Academic Press: New York, NY, USA, 1960; pp. 369–378. [Google Scholar]
- Castro-Acosta, M.L.; Lenihan-Geels, G.N.; Corpe, C.P.; Hall, W.L. Berries and anthocyanins: Promising functional food ingredients with postprandial glycaemia-lowering effects. Proc. Nutr. Soc. 2016, 75, 342–355. [Google Scholar] [CrossRef]
- Cao, H.; Ou, J.; Chen, L.; Zhang, Y.; Szkudelski, T.; Delmas, D.; Daglia, M.; Xiao, J. Dietary polyphenols and type 2 diabetes: Human Study and Clinical Trial. Crit. Rev. Food Sci. Nutr. 2019, 59, 3371–3379. [Google Scholar] [CrossRef]
- Alam, M.B.; An, H.; Ra, J.S.; Lim, J.Y.; Lee, S.H.; Yoo, C.Y.; Lee, S.-H. Gossypol from cottonseeds ameliorates glucose uptake by mimicking insulin signaling and improves glucose homeostasis in mice with streptozotocin-induced diabetes. Oxidative Med. Cell. Longev. 2018, 2018, 5796102. [Google Scholar] [CrossRef]
- Proença, C.; Freitas, M.; Ribeiro, D.; Oliveira, E.F.T.; Sousa, J.L.C.; Tomé, S.M.; Ramos, M.J.; Silva, A.M.S.; Fernandes, P.A. α-Glucosidase inhibition by flavonoids: An in vitro and in silico structure-activity relationship study. J. Enzym. Inhib. Med. Chem. 2017, 32, 1216–1228. [Google Scholar] [CrossRef] [Green Version]
- Yoon, E.K.; Jeong, Y.T.; Li, X.; Cui, S.; Park, D.C.; Kim, Y.H.; Kim, Y.D.; Chang, H.W.; Lee, S.-H.; Hwang, S.L. Glyceollin improves endoplasmic reticulum stress-induced insulin resistance through CaMKK-AMPK pathway in L6 myotubes. J. Nutr. Biochem. 2013, 24, 1053–1061. [Google Scholar] [CrossRef]
- Lee, D.S.; Lee, S.-H. Genistein, a soy isoflavone, is a potent alpha-glucosidase inhibitor. FEBS Lett. 2001, 501, 84–86. [Google Scholar] [CrossRef] [Green Version]
- Kazeem, M.I.; Adamson, J.O.; Ogunwande, I.A. Modes of inhibition of α-amylase and α-glucosidase by aqueous extract of Morinda lucida Benth leaf. Biomed. Res. Int. 2013, 2013, 527570. [Google Scholar] [CrossRef] [Green Version]
- Adisakwattana, S.; Chantarasinlapin, P.; Thammarat, H.; Yibchok-Anun, S. A series of cinnamic acid derivatives and their inhibitory activity on intestinal alpha-glucosidase. J. Enzym. Inhib. Med. Chem. 2010, 24, 1194–1200. [Google Scholar] [CrossRef]
- Thompson, W.J.; Appleman, M.M. Multiple cyclic nucleotide phosphodiesterase activities from rat brain. Biochemistry 1971, 10, 311–316. [Google Scholar]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Gallo-Cassarino, T.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014, 42, W252–W258. [Google Scholar] [CrossRef]
- Chou, T.-C. Theoretical basis, experiemntal design, and computerized simulation of synergism and antagonism in drug combination studies. Pharm. Rev. 2006, 58, 621–681. [Google Scholar] [CrossRef]
- Chiasson, J.L.; Josse, R.G.; Gomis, R.; Hanefeld, M.; Karasik, A.; Laakso, M. Acarbose for prevention of type 2 diabetes mellitus: The STOP-NIDDM randomised trial. Lancet 2002, 359, 2072–2077. [Google Scholar] [CrossRef]
- Wang, Y.; Ma, L.; Li, Z.; Du, Z.; Liu, Z.; Qin, J.; Wang, X.; Huang, Z.; Gu, L.; Chen, A.S. Synergetic inhibition of metal ions and genistein on α-glucosidase. FEBS Lett. 2004, 576, 46–50. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Ma, L.; Chen, W.H.; Park, H.; Ke, Z.; Wang, B. Binding mechanism and synergetic effects of xanthone derivatives as noncompetitive alpha-glucosidase inhibitors: A theoretical and experimental study. J. Phys. Chem. B 2013, 117, 13464–13471. [Google Scholar] [CrossRef]





| Inhibitor | Mode of Inhibition | Km (μm) | Ki (μm) |
|---|---|---|---|
| Acarbose | Competitive | 0.21 ± 0.02 | 45.88 ± 3.75 |
| Glyceollin | Competitive | 0.24 ± 0.03 | 18.99 ± 4.45 |
| Genistein | Noncompetitive | 0.36 ± 0.06 | 15.42 ± 2.48 |
| Luteolin | Mixed | 0.30 ± 0.06 | 16.81 ± 9.60 |
| Daidzein | Uncompetitive | 0.40 ± 0.05 | 9.99 ± 1.24 |
| IC50 | Mode of Inhibition | Substrate (PNPG, mM) | Remark | |||
|---|---|---|---|---|---|---|
| 0.125 | 0.25 | 0.5 | 1 | |||
| Glyceollin (μM) | Competitive | 16.57 ± 3.25 | 16.57 ± 0.15 | 19.11 ± 0.96 | 33.23 ± 3.97 | Synergistic |
| Luteolin (μM) | Mixed | 18.63 ± 1.20 | 14.82 ± 0.01 | 15.35 ± 1.99 | 19.52 ± 1.59 | |
| Glyceollin + Luteolin (μM) | 6.31 ± 0.59 *,# | 6.78 ± 0.18 *,# | 9.00 ± 0.40 *,# | 14.16 ± 0.35 * | ||
| Glyceollin (μM) | Competitive | 16.57 ± 3.25 | 16.57 ± 0.15 | 19.11 ± 0.96 | 33.23 ± 3.97 | Non-synergistic |
| Genistein (μM) | Noncompetitive | 40.47 ± 4.99 | 15.42 ± 0.33 | 12.00 ± 0.31 | 9.10 ± 0.61 | |
| Glyceollin + Genistein (μM) | 32.47 ± 1.48 | 33.73 ± 1.51 | 20.67 ± 0.44 | 21.81 ± 0.58 | ||
| Glyceollin (μM) | Competitive | 16.57 ± 3.25 | 16.57 ± 0.15 | 19.11 ± 0.96 | 33.23 ± 3.97 | Non-synergistic |
| Daidzein (μM) | Uncompetitive | 59.16 ± 6.10 | 27.56 ± 2.93 | 23.19 ± 1.04 | 20.53 ± 1.82 | |
| Glyceollin + Daidzein (μM) | 33.69 ± 0.65 | 22.78 ± 1.20 | 16.05 ± 0.51 | 19.04 ± 0.08 | ||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Son, H.-U.; Yoon, E.-K.; Yoo, C.-Y.; Park, C.-H.; Bae, M.-A.; Kim, T.-H.; Lee, C.H.; Lee, K.W.; Seo, H.; Kim, K.-J.; et al. Effects of Synergistic Inhibition on α-glucosidase by Phytoalexins in Soybeans. Biomolecules 2019, 9, 828. https://doi.org/10.3390/biom9120828
Son H-U, Yoon E-K, Yoo C-Y, Park C-H, Bae M-A, Kim T-H, Lee CH, Lee KW, Seo H, Kim K-J, et al. Effects of Synergistic Inhibition on α-glucosidase by Phytoalexins in Soybeans. Biomolecules. 2019; 9(12):828. https://doi.org/10.3390/biom9120828
Chicago/Turabian StyleSon, Hyeong-U, Eun-Kyeong Yoon, Chi-Yeol Yoo, Chul-Hong Park, Myung-Ae Bae, Tae-Ho Kim, Chang Hyung Lee, Ki Won Lee, Hogyun Seo, Kyung-Jin Kim, and et al. 2019. "Effects of Synergistic Inhibition on α-glucosidase by Phytoalexins in Soybeans" Biomolecules 9, no. 12: 828. https://doi.org/10.3390/biom9120828