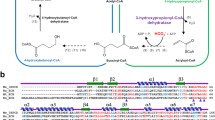
Abstract
Although enoyl-CoA hydratase/isomerase superfamily proteins are functionally diverse and extremely abundant in microbial and higher organism’s genome, they still have been elusively annotated. The genome of Cupriavidus necator H16 contains at least 54 enoyl-CoA hydratase/isomerase superfamily proteins that might influence on polyhydroxyalkanoate synthesis, but most of them are uncharacterized. Among them, we first determined crystal structure of H16_B0756 at a 2.0 Å resolution. The protein exhibits unique amino acid sequences compared to the other isoforms with identity lower than 36%. The structure of H16_B0756 forms a trimeric architecture and showed canonical disk-shape. Interestingly, H16_B0756 has only one glutamate residue at the active site while other enoyl-CoA hydratases have two nucleophilic glutamate at the catalytic site. We found that the active site conformation of H16_B0756 is quite similar to that of 1,2-epoxyphenylacetyl-CoA isomerase (PaaG) rather than those of other enoyl-CoA hydratases. In addition to the structural comparison, gene neighborhoods analysis suggested that H16_B0756 might function in the ring compound degradation.
Similar content being viewed by others
References
Mullernewen, G., U. Janssen, and W. Stoffel (1995) Enoyl-Coa Hydratase and Isomerase Form a Superfamily with a Common Active-Site Glutamate Residue. Eur. J. Biochem. 228: 68–73.
Xiang, H., L. S. Luo, K. L. Taylor, and D. Dunaway-Mariano (1999) Interchange of catalytic activity within the 2-enoylcoenzyme a hydratase isomerase superfamily based on a common active site template. Biochemistry-Us. 38: 7638–7652.
Chae, J. C., Y. Kim, Y. C. Kim, G. J. Zylstra, and C. K. Kim (2000) Genetic structure and functional implication of the fcb gene cluster for hydrolytic dechlorination of 4-chlorobenzoate from Pseudomonas sp DJ-12. Gene. 258: 109–116.
Holden, H. M., M. M. Benning, T. Haller, and J. A. Gerlt (2001) The crotonase superfamily: Divergently related enzymes that catalyze different reactions involving acyl coenzyme A thioesters. Accounts Chem. Res. 34: 145–157.
Mursula, A. M., D. M. F. van Aalten, J. K. Hiltunen, and R. K. Wierenga (2001) The crystal structure of Delta(3)-Delta(2)-enoyl-CoA isomerase. J. Mol. Biol. 309: 845–853.
Bahnson, B. J., V. E. Anderson, and G. A. Petsko (2002) Structural mechanism of enoyl-CoA hydratase: Three atoms from a single water are added in either an E1cb stepwise or concerted fashion. Biochemistry-Us. 41: 2621–2629.
Agnihotri, G., and H. W. Liu (2003) Enoyl-CoA hydratase: Reaction, mechanism, and inhibition. Bioorgan. Med. Chem. 11: 9–20.
Partanen, S. T., D. K. Novikov, A. N. Popov, A. M. Mursula, J. K. Hiltunen, and R. K. Wierenga (2004) The 1.3 angstrom crystal structure of human mitochondrial Delta(3)-Delta(2)-enoyl-CoA isomerase shows a novel mode of binding for the fatty acyl group. J. Mol. Biol. 342: 1197–1208.
Mursula, A. M., J. K. Hiltunen, and R. K. Wierenga (2004) Structural studies on Delta(3)-Delta(2)-enoyl-CoA isomerase: the variable mode of assembly of the trimeric disks of the crotonase superfamily. Febs. Lett. 557: 81–87.
Hamed, R. B., E. T. Batchelar, I. J. Clifton, and C. J. Schofield (2008) Mechanisms and structures of crotonase superfamily enzymes-How nature controls enolate and oxyanion reactivity. Cell Mol. Life Sci. 65: 2507–2527.
Geisbrecht, B. V., D. Zhu, K. Schulz, K. Nau, J. C. Morrell, M. Geraghty, H. Schulz, R. Erdmann, and S. J. Gould (1998) Molecular characterization of Saccharomyces cerevisiae Delta(3), Delta(2)-enoyl-CoA isomerase. J. Biol. Chem. 273: 33184–33191.
Geisbrecht, B. V., D. Y. Zhang, H. Schulz, and S. J. Gould (1999) Characterization of PECI, a novel monofunctional Delta(3), Delta(2)-enoyl-CoA isomerase of mammalian peroxisomes. J. Biol. Chem. 274: 21797–21803.
Choi, J. H., M. J. Seo, K. T. Lee, and D. K. Oh (2017) Biotransformation of Fatty Acid-rich Tree Oil Hydrolysates to Hydroxy Fatty Acid-rich Hydrolysates by Hydroxylases and their Feasibility as Biosurfactants. Biotechnol. Bioproc. E. 22: 709–716.
Gescher, J., W. Eisenreich, J. Worth, A. Bacher, and G. Fuchs (2005) Aerobic benzoyl-CoA catabolic pathway in Azoarcus evansii: studies on the non-oxygenolytic ring cleavage enzyme. Mol. Microbiol. 56: 1586–1600.
Gerratana, B., S. O. Arnett, A. Stapon, and C. A. Townsend (2004) Carboxymethylproline synthase from Pectobacterium carotorova: A multifaceted member of the Crotonase superfamily. Biochemistry-Us. 43: 15936–15945.
Haller, T., T. Buckel, J. Retey, and J. A. Gerlt (2000) Discovering new enzymes and metabolic pathways: Conversion of succinate to propionate by Escherichia coli. Biochemistry-Us. 39: 4622–4629.
Wong, B. J., and J. A. Gerlt (2004) Divergent function in the Crotonase superfamily: An anhydride intermediate in the reactions catalyzed by 3-hydroisobutyryl-CoA hydrolase (vol 125, pg 12076, 2003). J. Am. Chem. Soc. 126: 1921–1921.
Do, K. H., H. M. Park, S. K. Kim, and H. S. Yun (2018) Production of cis-Vaccenic Acid-oriented Unsaturated Fatty Acid in Escherichia coli. Biotechnol. Bioproc. E. 23: 100–107.
Truglio, J. J., K. Theis, Y. G. Feng, R. Gajda, C. Machutta, P. J. Tonge, and C. Kisker (2003) Crystal structure of Mycobacterium tuberculosis MenB, a key enzyme in vitamin K-2 biosynthesis. J. Biol. Chem. 278: 42352–42360.
Wong, B. J. and J. A. Gerlt (2004) Evolution of function in the crotonase superfamily: (3S)-methylglutaconyl-CoA hydratase from Pseudomonas putida. Biochemistry-Us. 43: 4646–4654.
Engemann, C., T. Elssner, S. Pfeifer, C. Krumbholz, T. Maier, and H. P. Kleber (2005) Identification and functional characterisation of genes and corresponding enzymes involved in carnitine metabolism of Proteus sp. Arch. Microbiol. 183: 176–189.
Patton, S. M., T. A. Cropp, and K. A. Reynolds (2000) A novel Delta(3),Delta(2)-enoyl-CoA isomerase involved in the biosynthesis of the cyclohexanecarboxylic acid-derived moiety of the polyketide ansatrienin A. Biochemistry-Us. 39: 7595–7604.
Gay, D. C., P. J. Spear, and A. T. Keatinge-Clay (2014) A Double-Hotdog with a New Trick: Structure and Mechanism of the trans-Acyltransferase Polyketide Synthase Enoyl-isomerase. Acs. Chem. Biol. 9: 2374–2381.
Lee, D. and K. J. Kim (2018) Structural Insight into Substrate Specificity of 3-Hydroxypropionyl-Coenzyme A Dehydratase from Metallosphaera sedula. Sci. Rep-Uk. 8.
Schwartz, E., B. Voigt, D. Zuhlke, A. Pohlmann, O. Lenz, D. Albrecht, A. Schwarze, Y. Kohlmann, C. Krause, M. Hecker, and B. Friedrich (2009) A proteomic view of the facultatively chemolithoautotrophic lifestyle of Ralstonia eutropha H16. Proteomics. 9: 5132–5142.
Chakraborty, P., K. Muthukumarappan, and W. R. Gibbons (2012) PHA Productivity and Yield of Ralstonia eutropha When Intermittently or Continuously Fed a Mixture of Short Chain Fatty Acids. J. Biomed. Biotechnol.
Eggers, J. and A. Steinbuchel (2013) Poly(3-Hydroxybutyrate) Degradation in Ralstonia eutropha H16 Is Mediated Stereoselectively to (S)-3-Hydroxybutyryl Coenzyme A (CoA) via Crotonyl-CoA. J Bacteriol. 195: 3213–3223.
Pohlmann, A., W. F. Fricke, F. Reinecke, B. Kusian, H. Liesegang, R. Cramm, T. Eitinger, C. Ewering, M. Potter, E. Schwartz, A. Strittmatter, I. Voss, G. Gottschalk, A. Steinbuchel, B. Friedrich, and B. Bowien (2006) Genome sequence of the bioplasticproducing “Knallgas” bacterium Ralstonia eutropha H16. Nat. Biotechnol. 24: 1257–1262.
Peplinski, K., A. Ehrenreich, C. Doring, M. Bomeke, and A. Steinbuchel (2010) Investigations on the microbial catabolism of the organic sulfur compounds TDP and DTDP in Ralstonia eutropha H16 employing DNA microarrays. Appl. Microbiol. Biot. 88: 1145–1159.
Volodina, E. and A. Steinbuchel (2014) (S)-3-hydroxyacyl-CoA dehydrogenase/enoyl-CoA hydratase (FadB') from fatty acid degradation operon of Ralstonia eutropha H16. Amb. Express. 4.
Seo, H. and K. Kim (2018) Purification, crystallization and X-ray crystallographic analysis of enoyl-CoA hydratase/isomerasefamily protein from Cupriavidus necator H16. Biodesign. 6: 46–49.
Park, S.-Y., S.-C. Ha, and Y.-G. Kim (2017) The protein crystallography beamlines at the pohang light source II. Biodesign. 5: 30–34.
Saitou, N. and M. Nei (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425.
Felsenstein, J. (1985) Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution. 39: 783–791.
Zuckerkandl, E. and L. Pauling (1965) Evolutionary divergence and convergence in proteins. pp. 97–166. Evolving genes and proteins. Elsevier, City.
Kumar, S., G. Stecher, M. Li, C. Knyaz, and K. Tamura (2018) MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 35: 1547–1549.
Holm, L. and L. M. Laakso (2016) Dali server update. Nucleic. Acids Res. 44: W351–W355.
Teufel, R., V. Mascaraque, W. Ismail, M. Voss, J. Perera, W. Eisenreich, W. Haehnel, and G. Fuchs (2010) Bacterial phenylalanine and phenylacetate catabolic pathway revealed. P Natl. Acad. Sci. USA. 107: 14390–14395.
Harwood, C. S. and R. E. Parales (1996) The beta-ketoadipate pathway and the biology of self-identity. Annu. Rev. Microbiol. 50: 553–590.
Brigham, C. J., D. R. Speth, C. Rha, and A. J. Sinskey (2012) Whole-Genome Microarray and Gene Deletion Studies Reveal Regulation of the Polyhydroxyalkanoate Production Cycle by the Stringent Response in Ralstonia eutropha H16. Appl. Environ. Microb. 78: 8033–8044.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Seo, H., Kim, KJ. Crystal Structure of a Novel Type Isomerase of Enoyl-CoA Hydratase/Isomerase Family Protein from Cupriavidus necator H16. Biotechnol Bioproc E 24, 155–162 (2019). https://doi.org/10.1007/s12257-018-0393-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12257-018-0393-3